Code
# Key Functions
st_filter(data_to_filter, reference_geometry, .predicate = relationship)
# .predicate = ""
st_intersects()
st_touches()
st_within()September 29, 2025
.shp (geometry – lat/long); .dbf (attribute); .sbx (binary spatial index file)Web Mercator (EPSG:3857): Web mapping standard, heavily distorts areas near poles
State Plane / UTM zones: local accuracy, different zones for different regions, optimized for specific geographic areas
Read Spatial Data: st_read
Spatial Subsetting: extract features based on spatial relationships
st_touches looks for neighborsst_withinst_intersects includes the reference feature itself.predicate specified, uses st_intersects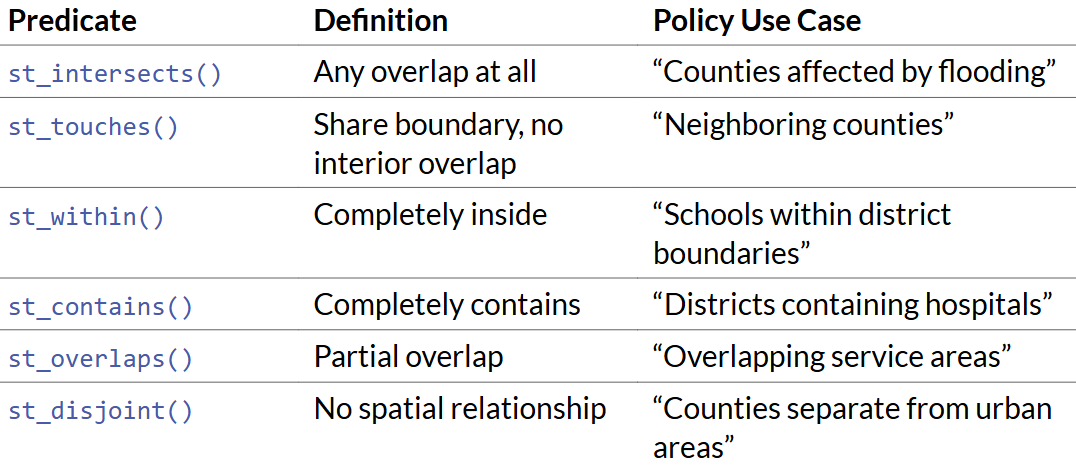
Checking and Setting CRS
st_crs(pa_counties)pa_counties <- st_set_crs(pa_counties, 4326)# st_transform
census_tracts <- census_tracts %>%
st_transform(st_crs(pa_counties))
# Transform to Pennsylvania South State Plane (good for PA analysis)
pa_counties_projected <- pa_counties %>% st_transform(crs = 3365)
# Transform to Albers Equal Area (good for area calculations)
pa_counties_albers <- pa_counties %>% st_transform(crs = 5070) `Plot Maps
Spatial Joins
Area Calculations
pa_countiesDistance and Buffers
# 10km buffer around all hospitals
hospitals_projected <- hospitals %>%
st_transform(crs = 3365)
st_crs(hospitals_projected)
hospital_buffers <- hospitals_projected %>%
st_buffer(dist = 32808.4) # 10,000 meters = 32808.4 feet
ggplot(hospital_buffers)+
geom_sf()+
theme_void()
# Different buffer sizes by hospital type (this is hypothetical, we don't have that column!)
hospital_buffers <- hospitals %>%
mutate(
buffer_size = case_when(
type == "Major Medical Center" ~ 15000,
type == "Community Hospital" ~ 10000,
type == "Clinic" ~ 5000
)
) %>%
st_buffer(dist = .$buffer_size)
# Use Unions to calculate what percentage of low-income tracts have access
access_summary <- low_income_tracts %>%
mutate(
has_access = st_intersects(., st_union(hospital_buffers), sparse = FALSE)
) %>%
st_drop_geometry() %>%
summarize(
total_tracts = n(),
tracts_with_access = sum(has_access),
pct_with_access = (tracts_with_access / total_tracts) * 100
)
print(access_summary)Load data → Get spatial boundaries and attribute data
Check projections → Transform to appropriate CRS
Join datasets → Combine spatial and non-spatial data
Spatial operations → Buffers, intersections, distance calculations
Aggregation → Summarize across spatial units
Visualization → Maps and charts
Interpretation → Policy recommendations
I was surprised at how intuitive and quick it was to develop maps and visualizations using packages in R as compared to ArcGIS. I discussed this with Prof. Delmelle during Office Hours, and she shared that she has relied exclusively on R for the past two years after switching to a Macbook.
While ArcGIS is still a useful tool and has many cool additional features such as StoryMaps or animations, I believe that R is the way forward for core data wrangling, visualization, and machine learning work.